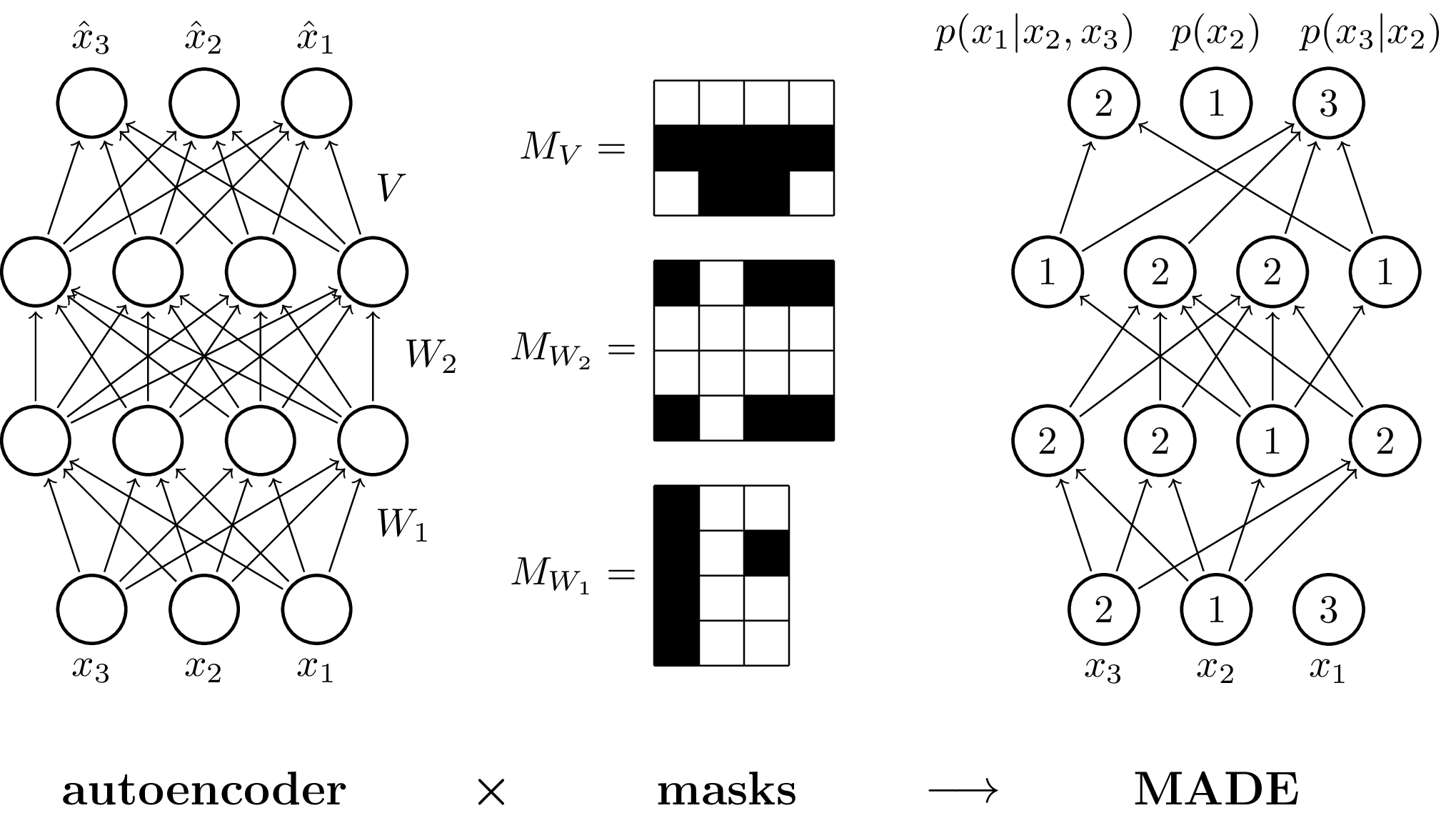
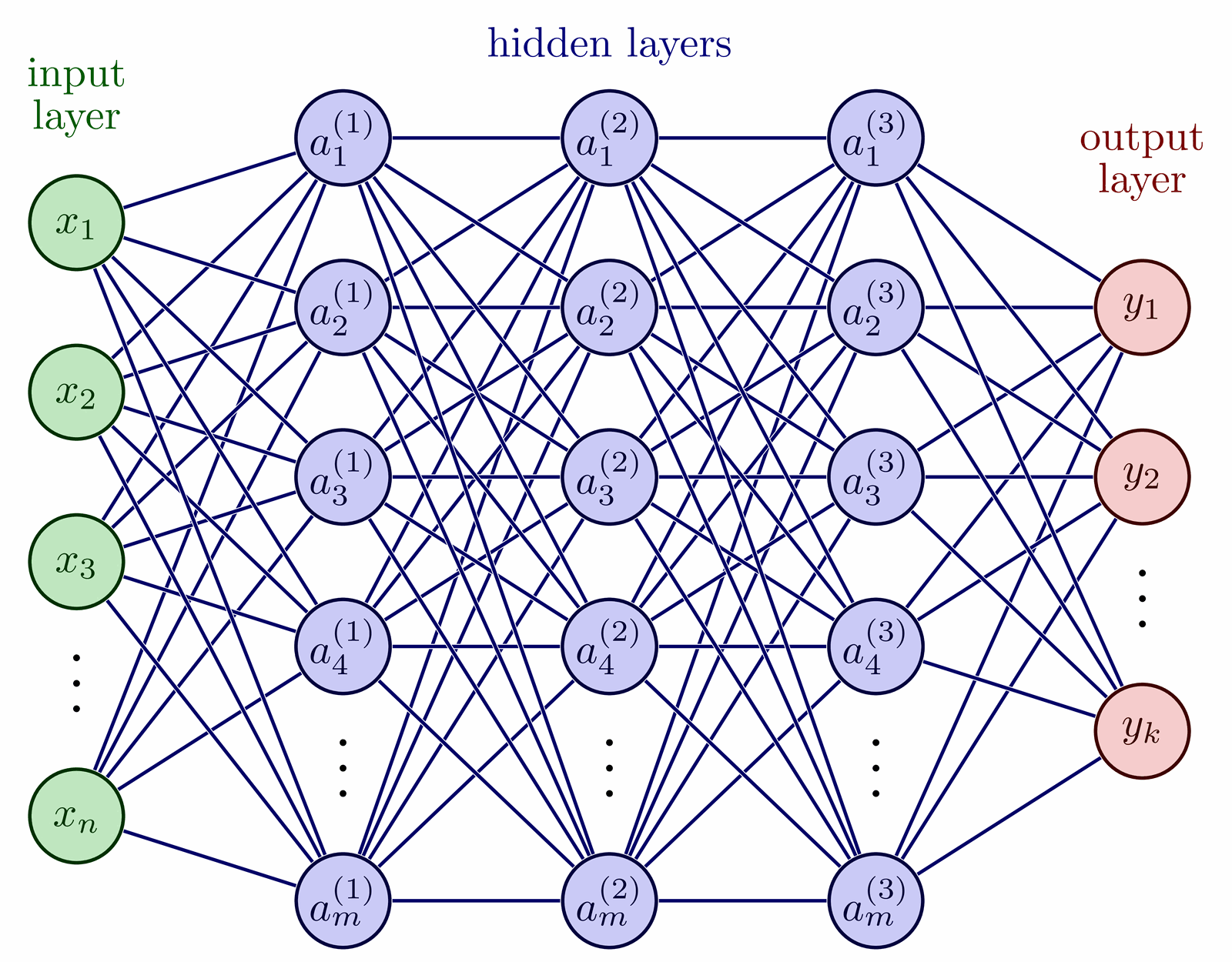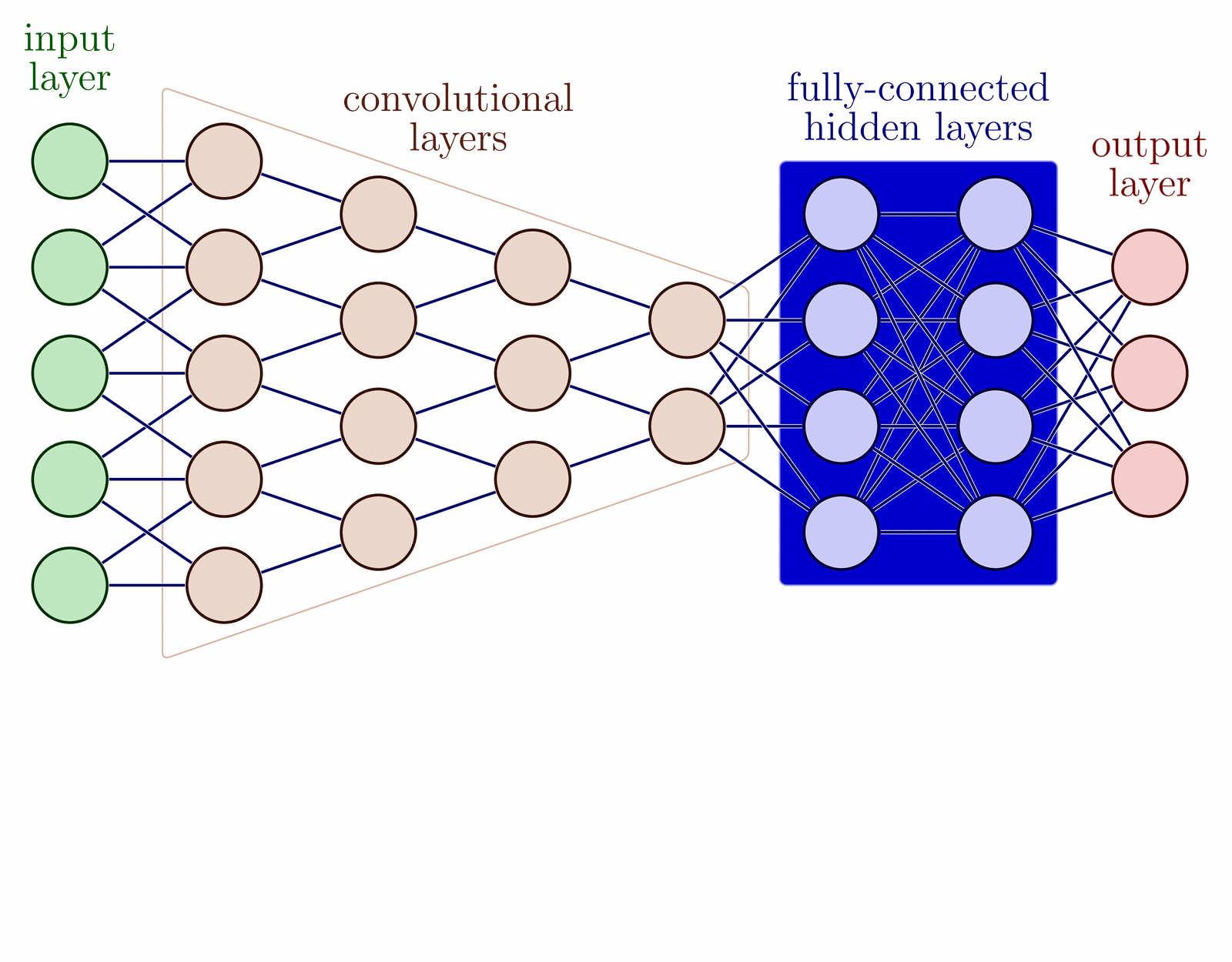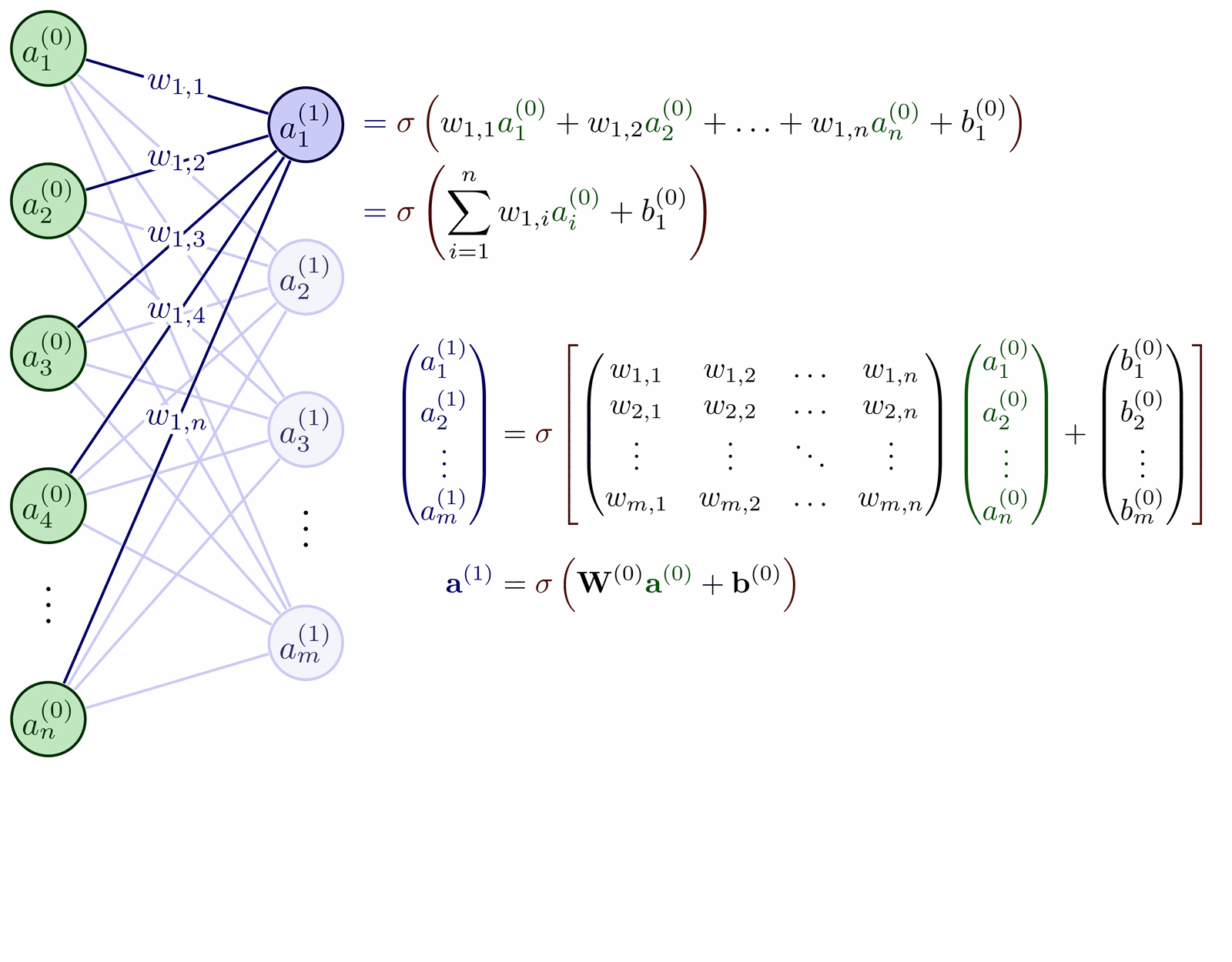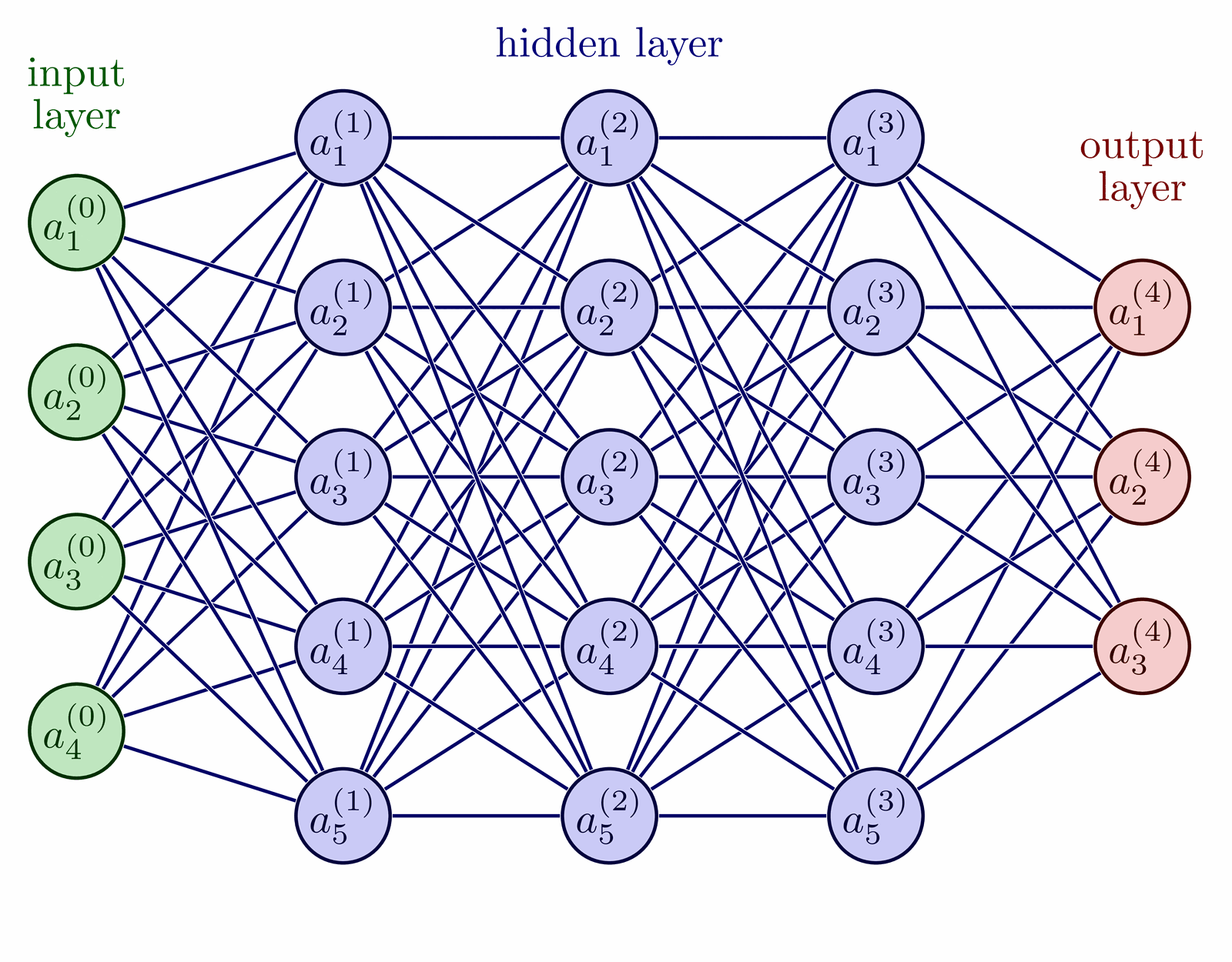
TikZ-reproduction of fig. 1 from the paper MADE: Masked Autoencoder for Distribution Estimation (arxiv:1502.03509).
Edit and compile if you like:
% TikZ-reproduction of fig. 1 from the paper MADE: Masked Autoencoder for Distribution Estimation (https://arxiv.org/abs/1502.03509).
\documentclass[tikz]{standalone}
\usepackage{xstring}
\usetikzlibrary{calc,positioning}
\newcommand\drawNodes[2]{
% #1 (str): namespace
% #2 (list[list[str]]): list of labels to print in the node of each neuron
\foreach \neurons [count=\lyrIdx] in #2 {
\StrCount{\neurons}{,}[\lyrLength] % use xstring package to save each layer size into \lyrLength macro
\foreach \n [count=\nIdx] in \neurons
\node[neuron] (#1-\lyrIdx-\nIdx) at (\lyrLength/2-\nIdx, 1.5*\lyrIdx) {\n};
}
}
\newcommand\denselyConnectNodes[2]{
% #1 (str): namespace
% #2 (list[int]): number of nodes in each layer
\foreach \n [count=\lyrIdx, remember=\lyrIdx as \previdx, remember=\n as \prevn] in #2 {
\foreach \y in {1,...,\n} {
\ifnum \lyrIdx > 1
\foreach \x in {1,...,\prevn}
\draw[->] (#1-\previdx-\x) -- (#1-\lyrIdx-\y);
\fi
}
}
}
\newcommand\connectSomeNodes[2]{
% #1 (str): namespace
% #2 (list[list[list[int]]]): for each node in each layer, list all connected nodes in the next layer
\foreach \layer [count=\lyrIdx, evaluate=\lyrIdx as \nextLyr using int(\lyrIdx+1)] in #2
\foreach \neuron [count=\nIdx] in \layer
\foreach \edge in \neuron
\draw[->] (#1-\lyrIdx-\nIdx) -- (#1-\nextLyr-\edge);
}
\begin{document}
\begin{tikzpicture}[
shorten >=1pt, shorten <=1pt,
neuron/.style={circle, draw, minimum size=4ex, thick},
legend/.style={font=\large\bfseries},
]
% Fully-connected neural net
\drawNodes{fcnn}{{{,,}, {,,,}, {,,,}, {,,}}}
\denselyConnectNodes{fcnn}{{3, 4, 4, 3}}
\path (fcnn-1-1) -- (fcnn-2-1) node[midway, right=1ex] (W1) {$W_1$};
\path (fcnn-2-1) -- (fcnn-3-1) node[midway, right=1ex] (W2) {$W_2$};
\path (fcnn-3-1) -- (fcnn-4-1) node[midway, right=1ex] (V) {$V$};
% MADE net
\begin{scope}[xshift=9cm]
\drawNodes{made}{{{3,1,2}, {2,1,2,2}, {1,2,2,1}, {3,1,2}}}
\connectSomeNodes{made}{{
{{}, {1,2,3,4}, {1,3,4}},
{{2,3}, {1,2,3,4}, {2,3}, {2,3}},
{{1,3}, {1}, {1}, {1,3}},
}}
\end{scope}
% Input + output labels
\foreach \idx in {1,2,3} {
\node[below=0 of fcnn-1-\idx] {$x_\idx$};
\node[above=0 of fcnn-4-\idx] {$\hat x_\idx$};
\node[below=0 of made-1-\idx] {$x_\idx$};
}
% MADE output labels
\node[xshift=2.5ex, above=0 of made-4-1] {$p(x_3|x_2)$};
\node[above=0 of made-4-2] {$p(x_2)$};
\node[xshift=-4ex, above=0 of made-4-3] {$p(x_1|x_2,x_3)$};
% Bottom legend
\node[legend, below=of fcnn-1-2] (encoder) {autoencoder};
\node[legend, below=of made-1-2] (made) {MADE};
\node[legend, right=2.5cm of encoder] (masks) {masks};
\node[legend, yshift=-1pt] (masks) at ($(encoder)!0.55!(masks)$) {\texttimes};
\node[legend, yshift=-1pt] (masks) at ($(masks)!0.65!(made)$) {$\longrightarrow$};
% Mask matrices
\begin{scope}[shift={(3cm,5cm)}, scale=0.4]
\draw (0,0) grid (4,3);
\node at (-1.8,1.5) {$M_V =$};
\fill[black] (0,1) rectangle ++(4,1);
\fill[black] (1,0) rectangle ++(2,1);
\begin{scope}[yshift=-5cm]
\draw (0,0) grid (4,4);
\node at (-1.8,2) {$M_{W_2} =$};
\fill[black] (0,0) rectangle ++(1,1);
\fill[black] (0,3) rectangle ++(1,1);
\fill[black] (2,0) rectangle ++(2,1);
\fill[black] (2,3) rectangle ++(2,1);
\end{scope}
\begin{scope}[yshift=-10cm]
\draw (0,0) grid (3,4);
\node at (-1.8,2) {$M_{W_1} =$};
\fill[black] (0,0) rectangle ++(1,4);
\fill[black] (2,2) rectangle ++(1,1);
\end{scope}
\end{scope}
\end{tikzpicture}
\end{document}
Click to download: made.tex
Open in Overleaf: made.tex
This file is available on tikz.netlify.app and on GitHub and is MIT licensed.
See more on the author page of Janosh Riebesell..