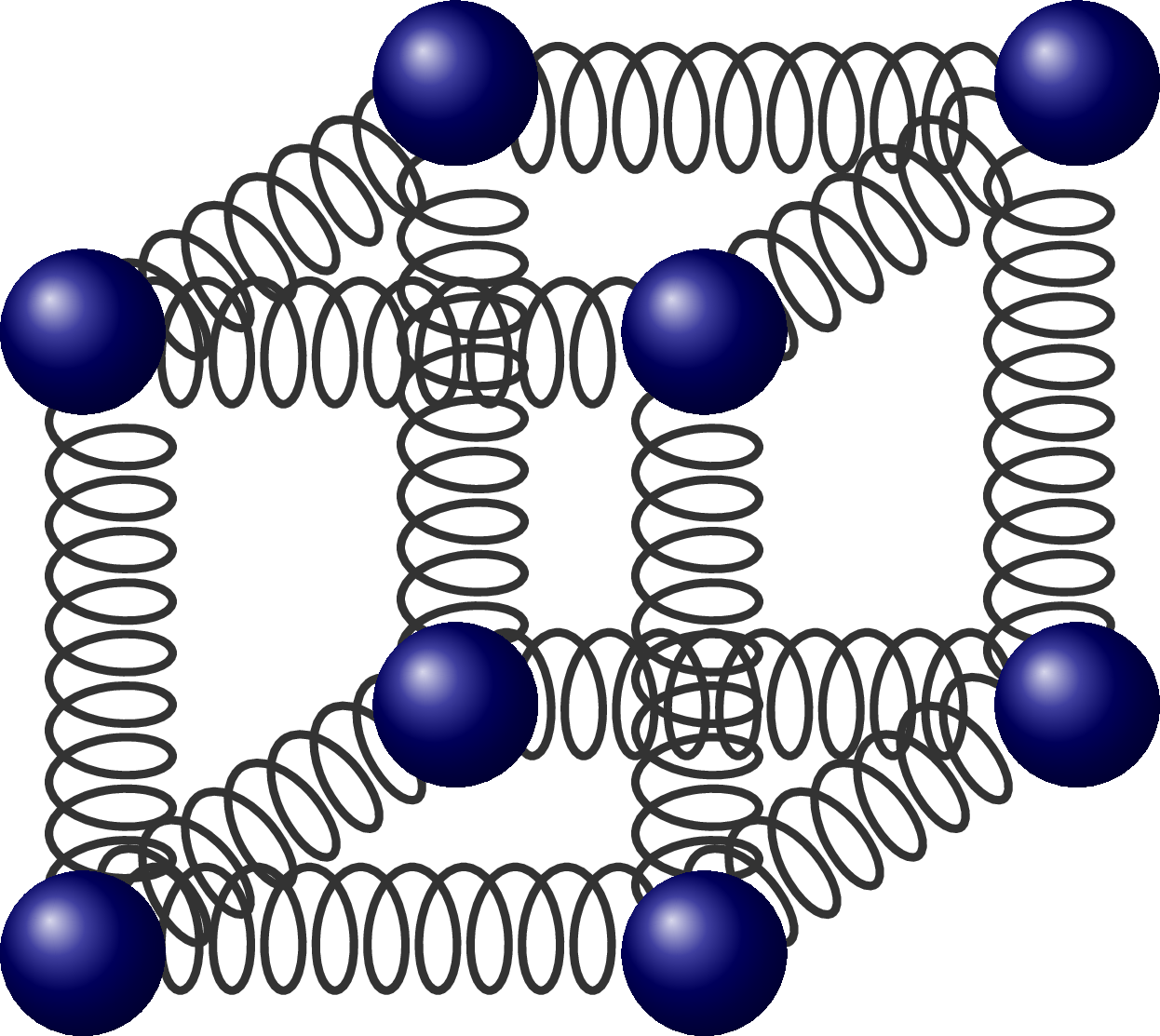
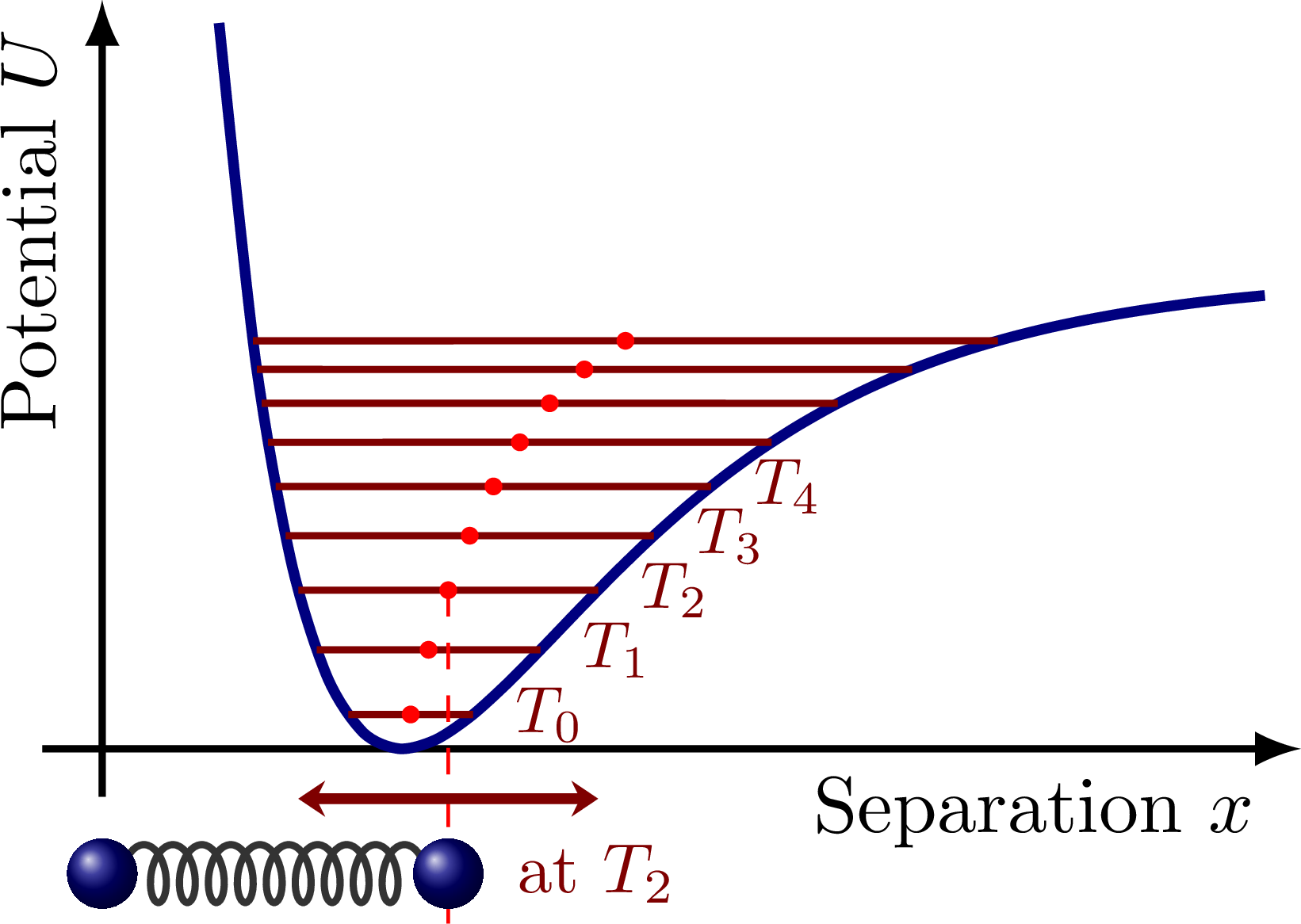
Molecular vibration illustrated with springs, and the Morse potential with different energy levels. Also see these diagrams on harmonic oscillator approximations.
Edit and compile if you like:
% Author: Izaak Neutelings (March 2019)
\documentclass[border=3pt,tikz]{standalone}
\usepackage{tikz}
\usepackage{ifthen}
\tikzset{>=latex} % for LaTeX arrow head
\usetikzlibrary{calc}
\usetikzlibrary{patterns,snakes,decorations.pathmorphing,intersections,arrows.meta}
\begin{document}
\tikzstyle{atom}=[ball color=blue!50!black]
\tikzstyle{bound}=[thick,black!80,decorate,decoration={coil,amplitude=6pt,segment length=5pt}]
% BOUNDS
\begin{tikzpicture}[scale=2,z={(0.6,0.4)}]
\def\z{-.03}
\def\o{1.03}
\draw[bound] (0,0,0) -- (1,0,0);
\draw[bound] (0,0,0) -- (0,1,0);
\draw[bound] (0,0,0) -- (0,0,1);
\draw[bound] (1,0,0) -- (1,1,0);
\draw[bound] (1,0,0) -- (1,0,1);
\draw[bound] (0,1,0) -- (1,1,0);
\draw[bound] (0,1,0) -- (0,1,1);
\draw[bound] (0,0,1) -- (0,1,1);
\draw[bound] (0,0,1) -- (1,0,1);
\draw[bound] (1,1,0) -- (1,1,1);
\draw[bound] (0,1,1) -- (1,1,1);
\draw[bound] (1,0,1) -- (1,1,1);
\fill[atom] (\z,\z,\z) circle (4pt);
\fill[atom] (\o,\z,\z) circle (4pt);
\fill[atom] (\z,\o,\z) circle (4pt);
\fill[atom] (\z,\z,\o) circle (4pt);
\fill[atom] (\o,\o,\z) circle (4pt);
\fill[atom] (\o,\z,\o) circle (4pt);
\fill[atom] (\z,\o,\o) circle (4pt);
\fill[atom] (\o,\o,\o) circle (4pt);
\end{tikzpicture}
% MORSE POTENTIAL
\begin{tikzpicture}
\def\A{1.9}
\def\a{1.1}
\def\r{1.2}
\def\m{1}
\def\N{30}
\def\xlow{{0.39*\r}}
\def\xmax{4.8}
\def\ymax{3.0}
\def\Emin{0.28}
\def\yA{-.5}
\def\E#1{{\Emin*(#1+1/2) - (\Emin*(#1+1/2))^2/(4*\A)}}
% AXIS
\draw[->,thick] (0,-0.04*\xmax) -- (0,\ymax)
node[left=2,anchor=south east,rotate=90,scale=0.9] {Potential $U$};
\draw[->,thick] (-0.05*\xmax,0) -- (\xmax,0)
node[left=2,anchor=north east,scale=0.9] {Separation $x$};
% MORSE POTENTIAL
\draw[very thick,blue!50!black,variable=\x,domain=\xlow:0.97*\xmax,samples=\N,smooth,name path=morse]
plot (\x,{\A*(1-exp(-\a*(\x-\r)))^2});
\path[name path=line1] (0,\E{0}) --++ (\xmax,0);
% ENERGY LEVEL
\foreach \i in {0,...,8}{
\path[name path=level\i] (0,\E{\i}) --++ (\xmax,0);
\draw[thick,red!50!black,name intersections={of=morse and level\i,name=E\i}]
(E\i-1) -- (E\i-2);
\ifthenelse{\i < 5}{ \node[right=2.2,red!50!black,scale=0.75] at (E\i-2) {$T_\i$}; }{}
\fill[red]
($(E\i-1)!0.5!(E\i-2)$) circle (1pt) coordinate (E\i);
}
% MOLECULE
\def\i{2}
\draw[dashed,red] (E\i) -- (E\i |- 0,\yA) coordinate (A\i) --++ (0,.4*\yA);
%node [below,red!90!black,scale=0.8] {$\langle r \rangle$};
\draw[thick,black!80,decorate,decoration={coil,amplitude=3.3pt,segment length=3.3pt}]
(0.08,\yA) -- ($(0,\yA)!0.95!(A\i)$);
\fill[atom] (0,\yA) circle (0.14);
\fill[atom] (A\i) circle (0.14)
node[right=5,red!50!black,scale=0.8] {at $T_\i$};
\draw[{Stealth[scale=1,length=3,width=4]}-{Stealth[scale=1,length=3,width=4]},very thick,red!50!black]
(E\i-1 |- 0,.4*\yA) -- (E\i-2 |- 0,.4*\yA);
\end{tikzpicture}
%% MORSE POTENTIAL (PGF)
%\begin{tikzpicture}
% \def\a{1}
% \def\r{1.5}
% \def\m{1}
% \def\N{30}
% \def\xmax{6}
% \def\ymax{1.5}
% \begin{axis}[every axis plot post/.append style={
% mark=none,domain={0.1*\r}:\xmax,samples=\N,smooth},
% xmin=(-.05*\xmax), xmax=(1.05*\xmax),
% ymin=(-.05*\ymax), ymax=(1.05*\ymax),
% restrict y to domain=0:{.9*\ymax},
% axis lines=middle,
% axis line style=thick,
% ticks=none,
% xlabel={intermolecular separation $r$},
% ylabel={potential $U$},
% xlabel style={anchor=north east},
% ylabel style={anchor=south east,rotate=90}, %at={(rel axis cs:-0.05,1)}
% width=9cm, height=7cm,
% clip mode=individual, %clip=false
% ]
%
% % MORSE POTENTIAL
% \addplot[very thick,blue!50!black,smooth] {(1-exp(-\a*(x-\r)))^2};
%
% %\clip (0.5,0.2) rectangle (2.5,1);
% \addplot[very thick,red!50!black] {0.5};
%
% % LABELS
% %\node[above=0pt,scale=0.75,red] at (1000,{planck(1000,3000)}) {\SI{3000}{K}};
%
% % MOLECULE
% %\draw[bound] (0.08,-0.2) -- (0.9*\r,-0.2);
% %\fill[atom] (0,-0.2) circle (6pt);
% %\fill[atom] (1,-0.2) circle (6pt);
%
% \end{axis}
%\end{tikzpicture}
\end{document}Click to download: molecules_vibrations.tex • molecules_vibrations.pdf
Open in Overleaf: molecules_vibrations.tex